Finding a Place in Open Source

This could be you! Click here to submit an abstract for our Maker Blog Series.
A considerable amount of life science research depends on the construction or analysis of recombinant DNA molecules. The exact sequence of these molecules is essential for reproducibility of the scientific work that they are a part of. An ad-hoc description of a cloning strategy unusually accompanies each publication. Unfortunately, most published cloning strategies are either incomplete or ambiguous, and some are plainly wrong. Manually following a cloning strategy is an arduous process, which may be why so many errors of this kind pass the peer review process. Pydna is an extension of Python for compact expression of cloning strategies. Pydna in a Jupyter Notebook can be read by users with rudimentary knowledge of Python, as the narrative format is similar to the traditional free-form description. These cloning strategies are easy to verify by simply re-executing the notebook. An added benefit is automatic verification using automatic integration services such as GitHub Actions.
Metabolic engineering is a life science field that often involves modulation of metabolic pathways for metabolite overproduction aiming to create so-called cell factories for biotechnologically interesting molecules. Cell factories are believed to play a future role in the transition to a green economy. In my own laboratory, we attempt to produce the fatty acid part of the capsaicin molecule that gives heat to the chili pepper by inserting new genes into baker’s yeast, essentially the same organism used to make bread at home.
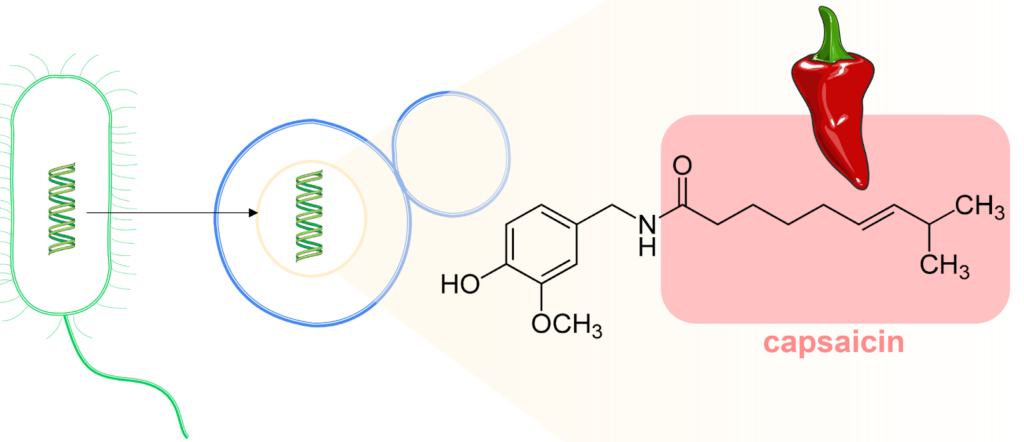
Figure 1: A gene from E. coli (green) is transferred to a Baker’s yeast cell (blue). The ultimate goal is to produce the fatty acid in capsaicin (red box). Figure by Cláudia Barata, University of Minho.
We need to make sure that new DNA molecules are stably maintained and functional within the cell by combining smaller DNA molecules into larger ones (recombinant DNA) according to a plan, usually referred to as a cloning strategy.
So why the rather provocative blog title? It turns out that many academic publications involving recombinant DNA do not contain a complete, unambiguous cloning strategy for the DNA constructions used in experiments. Cloning strategies are sometimes generic, like “gene X was cloned in plasmid Y resulting in plasmid Z.” (A plasmid is a small circular piece of DNA that can replicate on its own, and cloning in this context means simply joining DNA molecules together.) This is not nearly enough information to recreate plasmid Z, rendering the research on plasmid Z irreproducible. Failing to document the precise efforts that go into a cloning strategy is both unfortunate and unnecessary, as they consist of a series of simple unit operations strung together where the outcome is almost always deterministic.
Manual planning of cloning projects, where a point-and-click DNA editor is used together with sequence files obtained from databases or DNA sequencing experiments, is very common. These data files, sequence accession numbers, PCR primer sequences, etc. are needed alongside a detailed description of the manipulations performed in order to recreate the final construct sequence in-vitro or in-silico. Even when this information is provided, there is no guarantee that it is correct and complete unless one manually recreates each step. This may not be easy, however, as there is no widely-adopted standard for exactly what information should be included or how to describe a cloning strategy.
Cloning strategies can be formally expressed in Python using the pydna package. Pydna can handle the most common sub-cloning techniques (i.e. techniques for stitching DNA molecules together) in a compact manner. Jupyter Notebook supports the linear narrative format that is typically already used to describe cloning experiments.
One of several genetic modifications made in the context of synthesizing the capsaicin fatty acid in yeast is the expression of the fatty acid synthase gene AcpH (acyl carrier protein phosphodiesterase) from the bacteria E. coli. The cloning of this gene in a plasmid called pYPKa will be used as a simple example of how to use pydna.
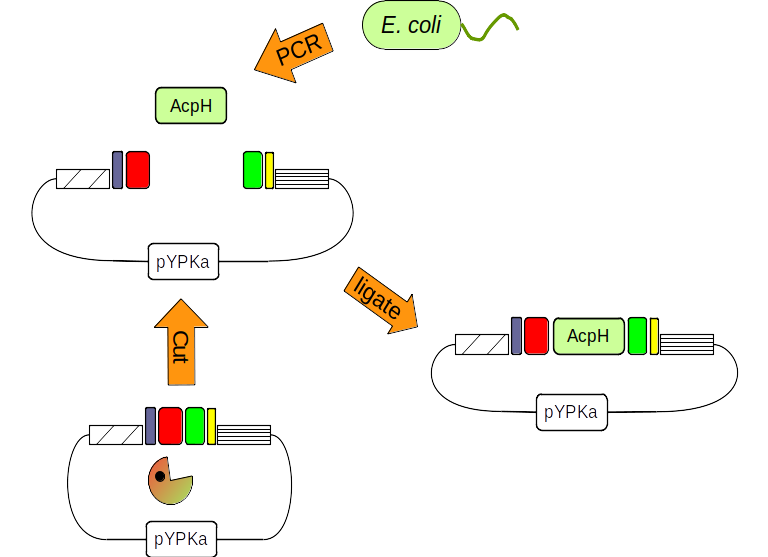
Figure 2: The pYPka plasmid is cut with a restriction enzyme (pacman). The AcpH gene from E. coli is amplified by PCR. Finally both DNA molecules are combined by ligation.
The first step is to import some needed functionality from pydna.

The pYPKa plasmid is read from a local text file. This DNA molecule is circular.

The restriction enzyme AjiI is imported from the Python package Biopython. (A restriction enzyme is a molecular scissor that can cut the DNA strands in a predictable manner.)

The circular plasmid is linearized using the restriction enzyme that cuts the plasmid in a single unique location.

Access to Genbank is needed in order to download the AcpH gene. The National Center for Biotechnology Information (NCBI) provides an API called “E-utilities” that can be accessed from Python. Change the email address below before executing this script as you should always provide an email address when using e-utilities.


The AcpH gene is downloaded from Genbank below.

The downloaded gene can be viewed here: NC_000913 424337-424918. We inspect the gene by printing it out the DNA sequence as a string:

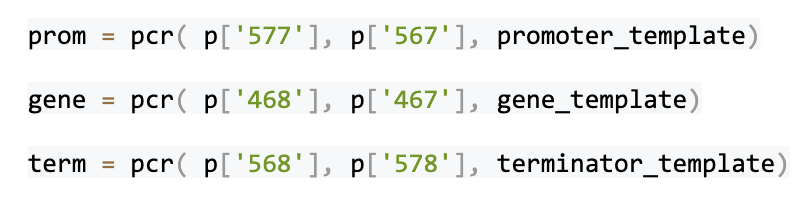
The gene is copied from the source DNA (often called a template DNA) by using the Polymerase Chain Reaction (PCR). A relatively short, specific DNA fragment is copied in large amounts relative to the template DNA. (PCR is incidentally also used for detecting infectious diseases like COVID-19.) The two PCR primers below (868 & 867) are used to PCR amplify the AcpH gene. PCR primers are short pieces of single-stranded DNA used to direct the PCR gene copying process.

The gene is synthesized by PCR using the two PCR primers:
We can visualize how the primers sit at each end of the AcpH gene like this:
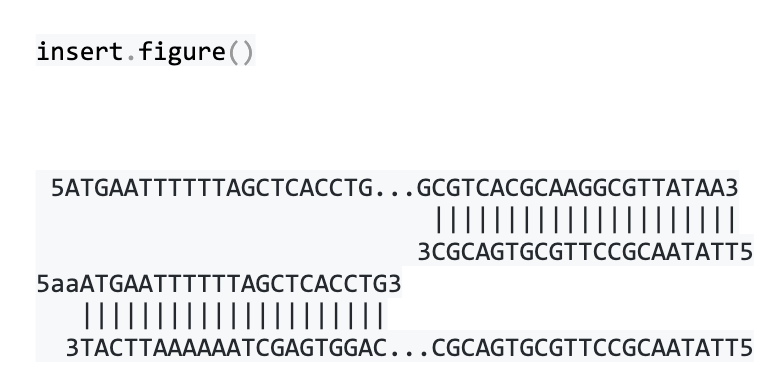
The final vector is ligated together from two linear DNA molecules:

We simply add the two linear DNA molecules together and tell pydna that we want a circular sequence using the looped method. We can then finally write the plasmid to a file using the write method.

A Jupyter notebook with the complete example above is available here.
The plasmid we made in the previous example is not sufficient for the gene to be active. The gene typically needs a promoter and a terminator in order to transcribe. Promoters and terminators are pieces of DNA that initiate and end transcription, which makes an RNA copy (mRNA) of the gene.
Homologous recombination and Gibson assembly techniques typically used in order to assemble larger, more complex recombinant DNA molecules. These techniques require short stretches of identical DNA sequences at the DNA ends. Pydna implements a DNA assembly algorithm that depends solely on the DNA sequence of the molecules. In the example below, we use pydna to simulate homologous recombination to make an expression plasmid for the gene we cloned in the previous example.
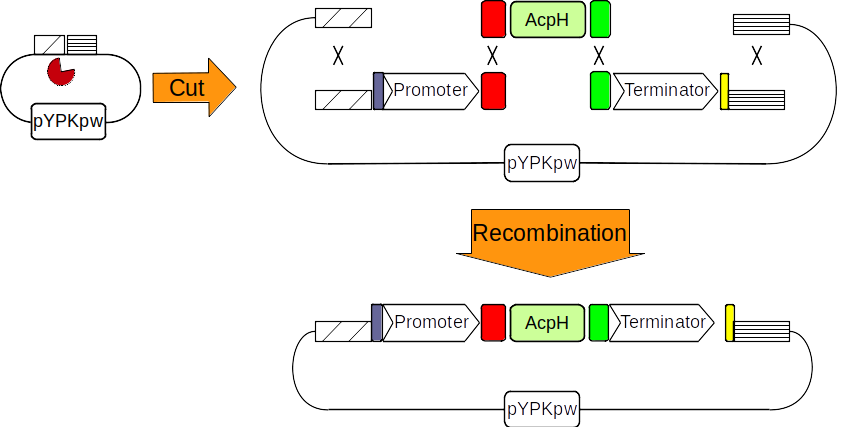
Figure 3: The pYPkpw plasmid is cut with a restriction enzyme (pacman). A promoter, the AcpH gene and a terminator are obtained through PCR. Four linear DNA molecules recombine together through shared DNA sequences to form a new circular molecule.
We read DNA sequences from local files.

The pYPKa_Z_RPL17A and pYPKa_E_RPL16B plasmids are similar to the plasmid constructed in the previous example, but carry a promoter and a terminator, respectively. We use three pairs of PCR primers to synthesize three PCR products:
A plasmid vector is linearized by digestion with the restriction enzyme EcoRV.

The assembly algorithm looks through the DNA sequences in the argument, discarding sequences shorter than a certain limit.

The assembly class has two methods to choose from, assemble_circular and assemble_linear, depending on the goal of the assembly. We are interested in circular assembly.

We got two circular products for the pYPK0_RPL17A_EcacpH_RPL16B vector. This is expected as the two molecules are each other’s reverse complement.

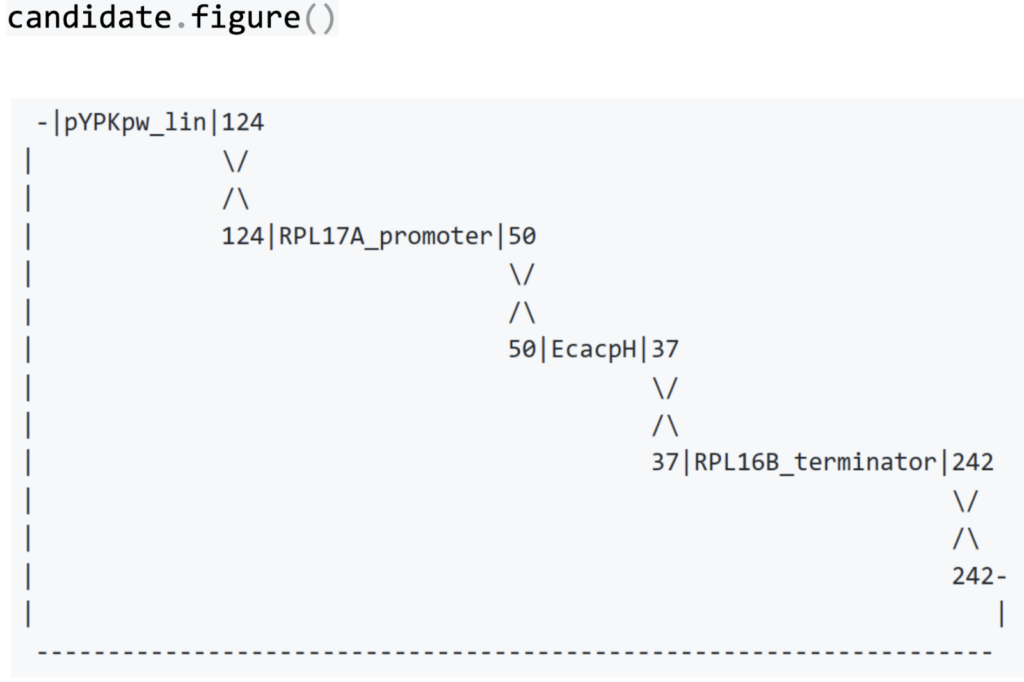
We can visualize how the four linear DNA molecules came together in a new circular plasmid. The numbers in the figure below are the lengths of the shared identical sequences.
The code examples above omit some essential steps for brevity. A Jupyter notebook with the complete homologous recombination example is available here.
A current barrier to the adoption of pydna is the slightly negative views associated with “writing code” still held by some biologists. However, programming will probably be an integral part of future biologists’ tool box due to the ever-increasing amounts of data generated in this field. Pydna and similar tools can help with the struggle for reproducibility in life science.
This work was supported by the Fundação para a Ciência e Tecnologia Portugal (FCT) through project FatVal PTDC/EAM-AMB/032506/2017 funded by national funds through the FCT I.P. and by the ERDF through the COMPETE2020 – Programa Operacional Competitividade e Internacionalizacão (POCI). CBMA was supported by the strategic program UIDB/04050/2020 funded by national funds through the FCT I.P.
Björn Johansson is an Assistant Professor in the Department of Biology at the University of Minho in Braga, Portugal, where he heads a small research group and teaches biology. His research area is yeast physiology and metabolic engineering, and his team is trying to broaden the metabolism of baker’s yeast to include interesting new products and substrates. Outside of biology, Björn is interested in bioinformatics and open reproducible science. He is also an open-source software enthusiast, using Linux on the desktop since 2007.
Anaconda is amplifying the voices of some of its most active and cherished community members in a monthly blog series. If you’re a Maker who has been looking for a chance to tell your story, elaborate on a favorite project, educate your peers, and build your personal brand, consider submitting an abstract. For more details and to access a wealth of educational data science resources and discussion threads—including one about this blog post—visit Anaconda Nucleus.
Talk to one of our experts to find solutions for your AI journey.